Parse raw plate reader data and convert it to a format compatible with QurvE
Source:R/data_parsers.R
parse_data.Rdparse_data takes a raw export file from a plate reader experiment (or similar device), extracts relevant information and parses it into the format required to run growth.workflow. If more than one read type is found the user is prompted to assign the correct reads to growth or fluorescence.
Usage
parse_data(
data.file = NULL,
map.file = NULL,
software = c("Gen5", "Gen6", "Biolector", "Chi.Bio", "GrowthProfiler", "Tecan",
"VictorNivo", "VictorX3"),
convert.time = NULL,
sheet.data = 1,
sheet.map = 1,
csvsep.data = ";",
dec.data = ".",
csvsep.map = ";",
dec.map = ".",
subtract.blank = TRUE,
calib.growth = NULL,
calib.fl = NULL,
calib.fl2 = NULL,
fl.normtype = c("growth", "fl2")
)Arguments
- data.file
(Character) A table file with extension '.xlsx', '.xls', '.csv', '.tsv', or '.txt' containing raw plate reader (or similar device) data.
- map.file
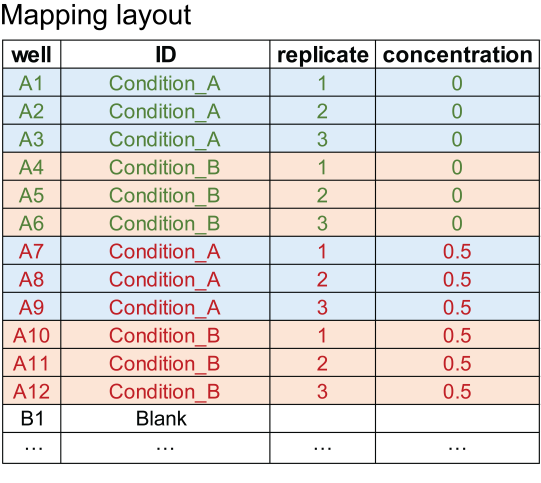
(Character) A table file in column format with extension '.xlsx', '.xls', '.csv', '.tsv', or '.txt' with 'well', 'ID', 'replicate', and 'concentration' in the first row. Used to assign sample information to wells in a plate.
- software
(Character) The name of the software/device used to export the plate reader data.
- convert.time
(
NULLor string) Convert time values with a formula provided in the form'y = function(x)'. For example:convert.time = 'y = 24 * x'- sheet.data, sheet.map
(Numeric or Character) Number or name of the sheets in XLS or XLSX files containing experimental data or mapping information, respectively (optional).
- csvsep.data, csvsep.map
(Character) separator used in CSV data files (ignored for other file types). Default:
";"- dec.data, dec.map
(Character) decimal separator used in CSV, TSV or TXT files with measurements and mapping information, respectively.
- subtract.blank
(Logical) Shall blank values be subtracted from values within the same experiment (TRUE, the default) or not (FALSE).
- calib.growth, calib.fl, calib.fl2
(Character or
NULL) Provide an equation in the form 'y = function(x)' (for example: 'y = x^2 * 0.3 - 0.5') to convert growth and fluorescence values. This can be used to, e.g., convert plate reader absorbance values into OD600 or fluorescence intensity into molecule concentrations. Caution!: When utilizing calibration, carefully consider whether or not blanks were subtracted to determine the calibration before selecting the inputsubtract.blank = TRUE.- fl.normtype
(Character string) Normalize fluorescence values by either diving by
'growth'or by fluorescence2 values ('fl2').
Value
A grodata object suitable to run growth.workflow. See read_data for its structure.
Examples
if(interactive()){
grodata <- parse_data(data.file = system.file("fluorescence_test_Gen5.xlsx", package = "QurvE"),
sheet.data = 1,
map.file = system.file("fluorescence_test_Gen5.xlsx", package = "QurvE"),
sheet.map = "mapping",
software = "Gen5",
convert.time = "y = x * 24", # convert days to hours
calib.growth = "y = x * 3.058") # convert absorbance to OD values
}