Generic plot function for flcFittedLinear objects. Plot the results of a linear regression on ln-transformed data
Source: R/fluorescence_plots.R
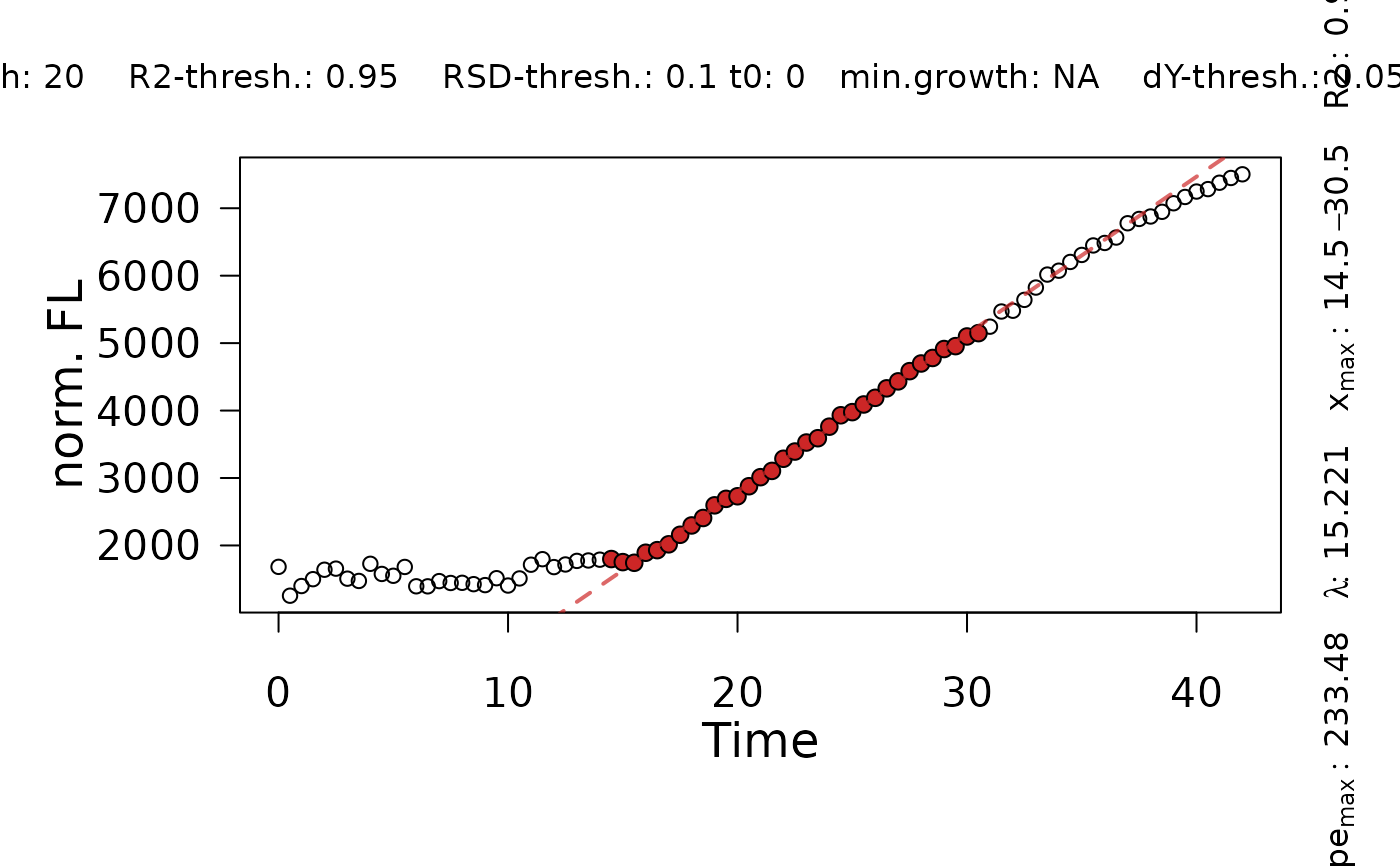
plot.flFitLinear.Rdplot.flFitLinear shows the results of a linear regression and visualizes raw data, data points included in the fit, the tangent obtained by linear regression, and the lag time.
Usage
# S3 method for flFitLinear
plot(
x,
log = "",
which = c("fit", "diagnostics", "fit_diagnostics"),
pch = 21,
cex.point = 1,
cex.lab = 1.5,
cex.axis = 1.3,
lwd = 2,
color = "firebrick3",
y.lim = NULL,
x.lim = NULL,
plot = TRUE,
export = FALSE,
height = ifelse(which == "fit", 7, 5),
width = ifelse(which == "fit", 9, 9),
out.dir = NULL,
...
)Arguments
- x
A
flFittedLinearobject created withflFitLinearor stored within aflFitResorflFitobject created withfl.workfloworflFit, respectively.- log
("x" or "y") Display the x- or y-axis on a logarithmic scale.
- which
("fit" or "diagnostics") Display either the results of the linear fit on the raw data or statistical evaluation of the linear regression.
- pch
(Numeric) Shape of the raw data symbols.
- cex.point
(Numeric) Size of the raw data points.
- cex.lab
(Numeric) Font size of axis titles.
- cex.axis
(Numeric) Font size of axis annotations.
- lwd
(Numeric) Line width.
- color
(Character string) Enter color either by name (e.g., red, blue, coral3) or via their hexadecimal code (e.g., #AE4371, #CCFF00FF, #0066FFFF). A full list of colors available by name can be found at http://www.stat.columbia.edu/~tzheng/files/Rcolor.pdf
- y.lim
(Numeric vector with two elements) Optional: Provide the lower (
l) and upper (u) bounds on y-axis as a vector in the formc(l, u).- x.lim
(Numeric vector with two elements) Optional: Provide the lower (
l) and upper (u) bounds on the x-axis as a vector in the formc(l, u).- plot
(Logical) Show the generated plot in the
Plotspane (TRUE) or not (FALSE).- export
(Logical) Export the generated plot as PDF and PNG files (
TRUE) or not (FALSE).- height
(Numeric) Height of the exported image in inches.
- width
(Numeric) Width of the exported image in inches.
- out.dir
(Character) Name or path to a folder in which the exported files are stored. If
NULL, a "Plots" folder is created in the current working directory to store the files in.- ...
Further arguments to refine the generated base R plot.
Examples
# load example dataset
input <- read_data(data.growth = system.file("lac_promoters.xlsx", package = "QurvE"),
data.fl = system.file("lac_promoters.xlsx", package = "QurvE"),
sheet.growth = 1,
sheet.fl = 2 )
#> Sample data are stored in columns. If they are stored in row format, please run read_data() with data.format = 'row'.
# Extract time and normalized fluorescence data for single sample
time <- input$time[4,]
data <- input$norm.fluorescence[4,-(1:3)] # Remove identifier columns
# Perform linear fit
TestFit <- flFitLinear(time = time,
fl_data = data,
ID = "TestFit",
control = fl.control(fit.opt = "l", x_type = "time",
lin.R2 = 0.95, lin.RSD = 0.1,
lin.h = 20))
plot(TestFit)